Abstract
Introduction:
Genetic profiling of AML reveals distinct molecular subgroups that inform disease classification and prognostic stratification (Tayyab M et al., J. Cancer Sci. Ther. 2014). Studies have demonstrated, that the Isocitrate Dehydrogenase Isoenzymes (IDH1/2), regulating cellular metabolism co-operate to result in AML (Papaemmanuil E et al., N. Engl. J. Med. 2016). Both IDH1/2 mutations produce neomorphic activity resulting in the overproduction of an onco-metabolite 2-hydroxyglutarate (2HG) (Gross S et al., J Exp Med. 2010). IDH1 mutations affect codon 132 and IDH2 mutations affect codon 140 or 172. IDH1 mutations have been reported to occur in 6-9% of adult and 1% of pediatric AML cases respectively, while IDH2 mutations have been reported to occur in 8-12% of adult and 1-2% of pediatric AML cases respectively (Abbas, S, et al., Blood. 2010; Andersson AK et al., Leukemia. 2011). The objective of this study was to conduct workflow analysis of the Abbott RealTime IDH1 and IDH2 assays in comparison to the Sanger based sequencing assays for IDH1/2 in blood/bone marrow specimens from subjects with advanced hematological malignancies. Next Generation Sequencing (NGS) was utilized to resolve samples with discordant results.
Methods:
This study was approved by Medical College of Wisconsin Institutional Review Board. The clinical specimens used were blinded during the study. Blood or bone marrow aspirates from 100 AML subjects sent to the BloodCenter of Wisconsin (BCW) were tested for IDH1/2 with four different assays (Abbott RealTime IDH1, Abbott RealTime IDH2 assays, the BCW's Polymerase Chain Reaction (PCR) based Sanger sequencing assay and NGS HemeOnc panel (contains 30 genes including IDH1/2). Abbott RealTime IDH1 and IDH2 assays are qualitative in vitro PCR tests wherein, the IDH1 assay detects five IDH1 mutations (R132H, R132C, R132L, R132G and R132S) and the IDH2 assay detects nine IDH2 mutations (R140Q, R140L, R140G, R140W, R172K, R172M, R172G, R172S and R172W).
The BCW's PCR based Sanger sequencing assay and NGS HemeOnc panel detect mutations in exon 4 of IDH1/2 with a sensitivity limit of 20% and 10% respectively. In this study NGS data was analyzed for only IDH1 and IDH2 .
DNA was extracted from 100 clinical specimens either by the Abbott m Sample preparation system or by the Qiagen DNA extraction method and tested using the Abbott RealTime IDH1 and IDH2 assays. Additionally, a set of commercially available controls (IDH1 R132H/ IDH2 R172K, IDH1 R132C/ IDH2 R140Q) with different mutation levels (20%, 5%, 1% and 0.25%) were tested using the above mentioned four assays
Results:
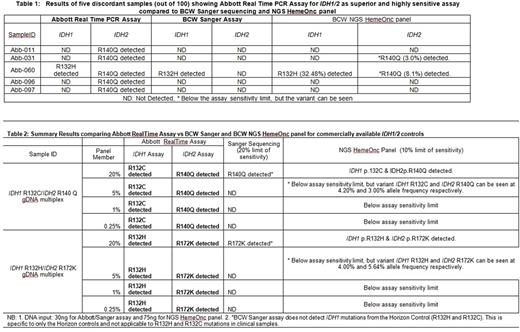
The Abbott RealTime IDH1 and IDH2 assays demonstrated 100% sensitivity and 95% specificity compared to the BCW Sanger sequencing assay. Five out of 100 samples with low level IDH2 mutations were missed by BCW Sanger sequencing assay but were detected as mutation positive by Abbott RealTime IDH2 assay. These five discordant samples with low level IDH2 mutations were subsequently tested with the BCW's NGS assay and two of the five samples were detected as positive by the NGS assay (Table 1). Additionally, turnaround time (TAT) analysis demonstrated that the results from the Abbott RealTime IDH1/2 assays were available in 2 business days as compared to 10 days for the BCW Sanger sequencing assay and 21 days for the BCW NGS HemOnc panel.
Testing of the mixture of IDH1 and IDH2 mutations with differing mutation levels (20%, 5%, 1% and 0.25%) resulted in the Abbott RealTime IDH1 and IDH2 assay reliably detecting the mutations down to the 1% level (with some variability at 0.25%) (Table2).
Conclusions:
This study demonstrated that the Abbott RealTime IDH1 and IDH2 assays detected IDH1 and IDH2 mutations at 1% mutation level in AML patients. The Abbott assays were highly sensitive (1%) compared to the PCR based Sanger sequencing and NGS HemeOnc panel, which have sensitivity limits of 20% and 10% respectively. The higher sensitivity and relatively fast TAT of the Abbott assays may potentially improve the ability to diagnose new and relapsed patients in settings of lower disease burden. The relatively fast TAT could facilitate clinicians in making a quicker decision about therapeutic treatment recommendations for afflicted patients.
Dash: Abbott Molecular Inc.: Other: I do not have financial relationship, however wanted to mention here is that this study was sponsored by Abbott Molecular Inc which was performed in BloodCenter of Wisconsin.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal